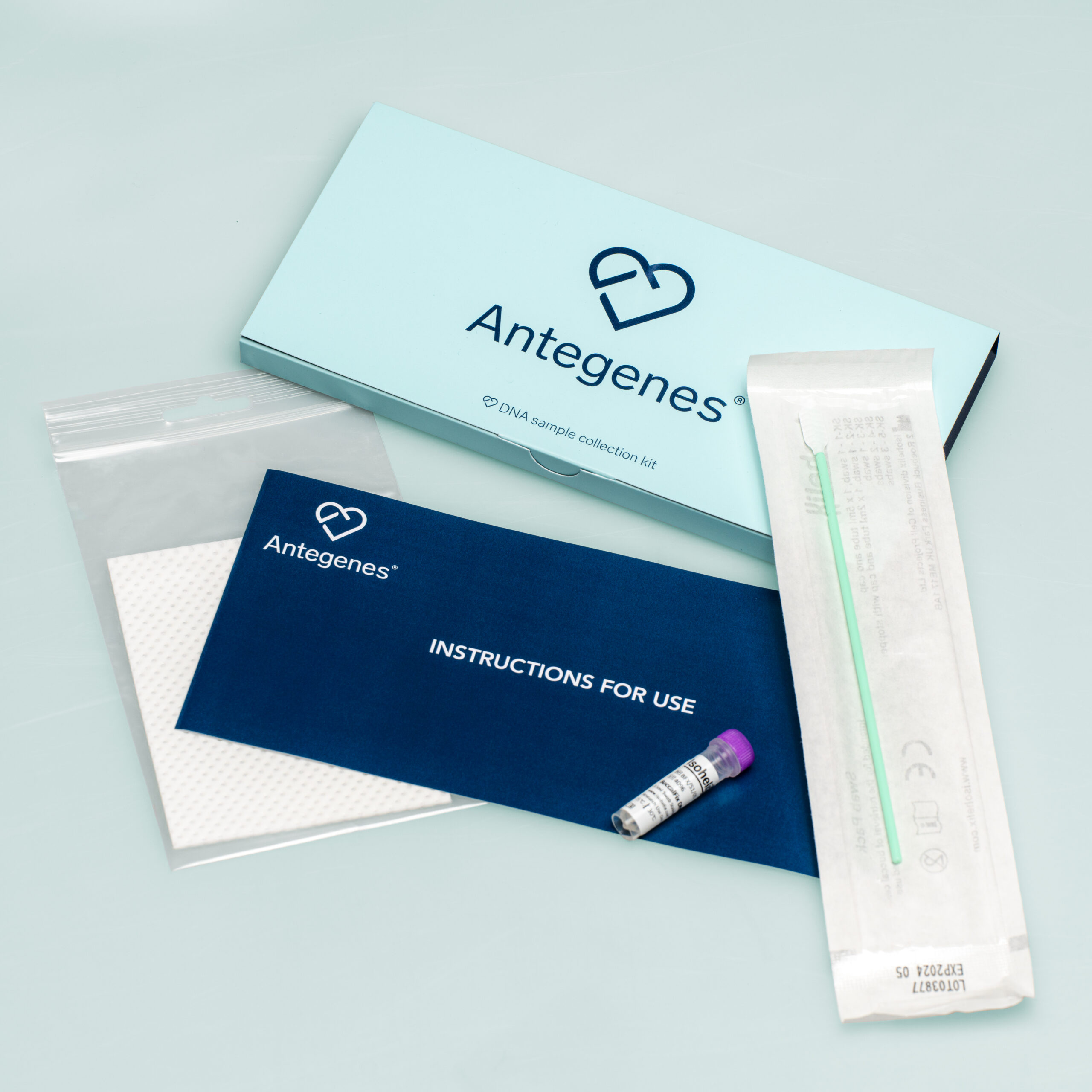
Personalised cancer prevention is an up-to-date solution aimed at minimising the potential impact of three different cancers by tailoring it to your genetic risk level.
Tests take into account your unique characteristics – genetic composition, sex, age and ethnicity – which make your results fully personalised.